Welcome to Vivarium E. coli’s documentation!
Vivarium E. coli project is a port of the Covert Lab’s E. coli Whole Cell Model to the Vivarium framework.
Warning
This documentation is very much a work in progress. It likely contains errors and poor formatting.
The scope of this project is to migrate the Whole Cell Model’s processes, and therefore takes the model’s sim_data as its starting point in the simulation pipeline. sim_data is a large configuration object created by the parameter calculator (ParCa). For this reason the reconstruction/ and wholecell/utils/ folders have been duplicated here as they are necessary to unpickle the serialized sim_data object. If a new sim_data object is required to be read, the corresponding wcEcoli folders will have to be synchronized.
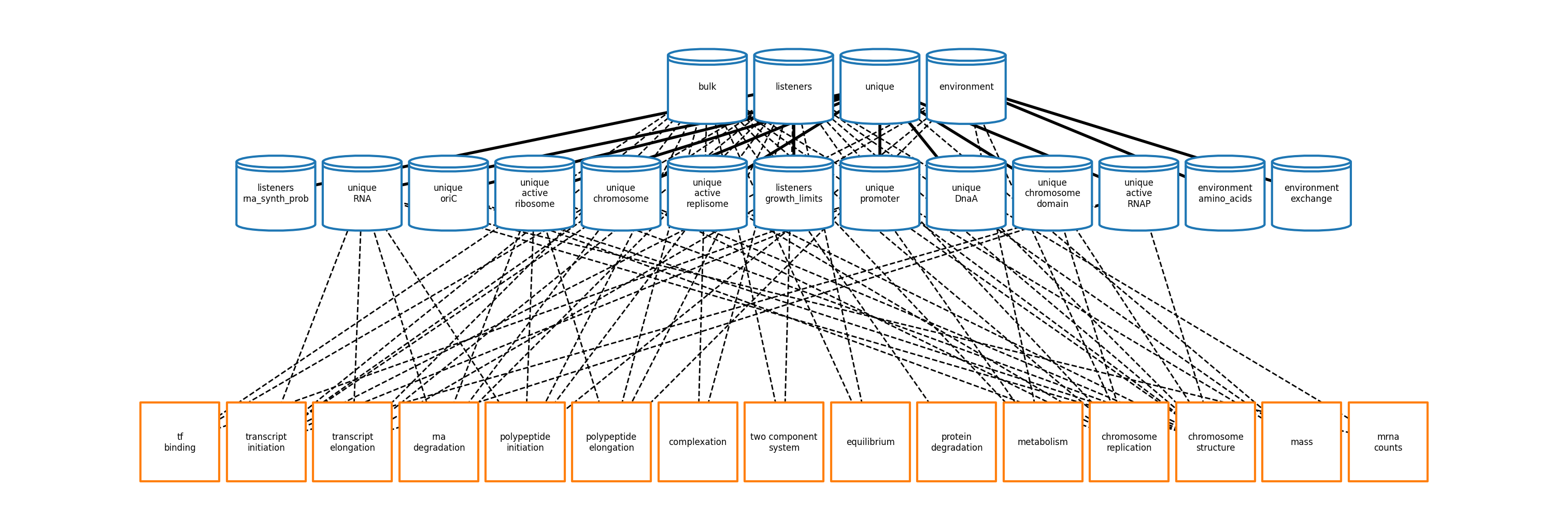
All state handling (previously handled by Bulk- and UniqueMolecules states/containers/views) and the actual running of the simulation (previously wholecell.sim.Simulation) are now handled entirely by Vivarium’s core engine and process interface.
The new process classes can be found in ecoli/processes/* and are linked together using a Vivarium topology that is generated in ecoli/experiments/ecoli_master_sim.py.