Vivarium E. coli Processes
This notebook demonstrates how the processes work individually. For each process, we will:
Load in the process and parameters
Plot a toplogy diagram
The topology is a network that demonstrates how a process connects to its stores (which hold state variables).
Display the ports schema
The port schema defines a systems ports (top-level keys), and the expected behavior of molecules under that port (its schema)
*is a wild card, specifies the schema of everything that can go into the port
Simulate the process
Demonstrate distinct features of that process
Miscellaneous notes/ideas: - consider adding interactive widgets (plotly) - users can click boxes to choose which molecules to plot
[1]:
# Make sure notebook runs out of vivarium-ecoli directory
import sys, os
# get the path to the notebook, and change working directory
notebook_path = sys.path[0][:sys.path[0].index('notebooks')]
sys.path.append(notebook_path)
os.chdir(sys.path[-1])
cwd = os.getcwd()
Load the required components
import modules
[2]:
from vivarium.core.process import Process
from vivarium.core.store import Store
from vivarium.core.engine import Engine, pp
from vivarium.core.composition import simulate_process, simulate_composite
from vivarium.plots.topology import plot_topology
from vivarium.plots.simulation_output import plot_variables
from ecoli.processes.registries import topology_registry
import ecoli
import copy
Load sim_data
This unpickles and sim_data object, which holds model parameters from wcEcoli. LoadSimData includes functions to retrieve individual processes’ parameters, which can be modified and passed into their respective process models.
[3]:
from ecoli.library.sim_data import LoadSimData
SIM_DATA_PATH = 'reconstruction/sim_data/kb/simData.cPickle'
load_sim_data = LoadSimData(
sim_data_path=SIM_DATA_PATH,
seed=0)
Get initial state snapshot
initial_state is a dict with the initial state of the system – a snapshot saved from wcEcoli.
[4]:
from ecoli.composites.ecoli_master import get_state_from_file
INITIAL_STATE_PATH = 'data/wcecoli_t1000.json'
initial_state = get_state_from_file(path=INITIAL_STATE_PATH)
Helper functions for executing the notebook
[5]:
schema_keys = Store.schema_keys
def make_port_printout(ports_schema, depth=0, schema_show=5, filler_size=5):
print_dict = ''
filler = filler_size * ' '
for port, schema in ports_schema.items():
if isinstance(schema, dict):
schemavars = list(schema.keys())
if any(var in schemavars for var in schema_keys):
print_schema = ''
for k, v in schema.items():
print_schema += f'{(depth+1) * filler} {k}: {v}\n'
print_dict += f'{depth * filler}{port}:\n{print_schema}\n'
else:
schema_items = schema.items()
first_schema = dict(list(schema_items)[:schema_show])
next_print = make_port_printout(first_schema, depth+1)
print_dict += f'{port}:\n{next_print}\n'
if len(schema) > schema_show:
print_dict += f'{(depth+1) * filler}'
print_dict += f'... skipping {len(schema)-schema_show} schema entries ...'
print_dict += f'\n\n'
else:
print_dict += f'{filler}{schema}\n'
return print_dict
def find_increasing(d):
for key, value in d.items():
if value[-1] > value[0]:
return {key: value}
def find_decreasing(d):
for key, value in d.items():
if value[-1] < value[0]:
return {key: value}
Complexation
[6]:
from ecoli.processes.complexation import Complexation
# load in parameters
cplx_config = load_sim_data.get_complexation_config()
cplx_config['time_step'] = 0.01
# initialize process
complexation = Complexation(cplx_config)
[7]:
# plot topology
cplx_topology_plot_settings = {
'buffer': 1,
'node_labels': {
'ecoli-complexation': 'ecoli\ncomplexation'
},
'show_ports': False,
'node_size': 10000,
'dashed_edges': True,
'node_distance': 4,
}
cplx_topology_fig = plot_topology(complexation, cplx_topology_plot_settings)
[8]:
# display ports schema
cplx_ports = complexation.ports_schema()
cplx_printout = make_port_printout(cplx_ports)
print(cplx_printout)
molecules:
1-PFK[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
1-PFK-MONOMER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
2OXOGLUTARATEDEH-CPLX[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
E1O[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
E2O[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
... skipping 2569 schema entries ...
listeners:
complexation_events:
_default: []
_updater: set
_emit: True
[9]:
# tweak initial state
cplx_initial_state = copy.deepcopy(initial_state)
cplx_initial_state['bulk']['6PGLUCONDEHYDROG-MONOMER[c]'] = 1000
# run simulation and retrieve final data
cplx_settings = {
'total_time': 0.1,
'initial_state': cplx_initial_state,
'topology': complexation.topology}
cplx_data = simulate_process(complexation, cplx_settings)
Simulation ID: a7b0130c-2025-11ec-8f9d-8c85908ac627
Created: 09/27/2021 at 23:31:04
Completed in 0.178275 seconds
For complexation, let’s look at the 6PGLUCONDEHYDROG-MONOMER[c] monomer as it transitions to the 6PGLUCONDEHYDROG-CPLX[c] complex:
[10]:
# plot output
cplx_fig = plot_variables(
cplx_data,
variables=[
('bulk', '6PGLUCONDEHYDROG-MONOMER[c]'),
('bulk', '6PGLUCONDEHYDROG-CPLX[c]'),
],
column_width=10, row_height=3, row_padding=0.5)
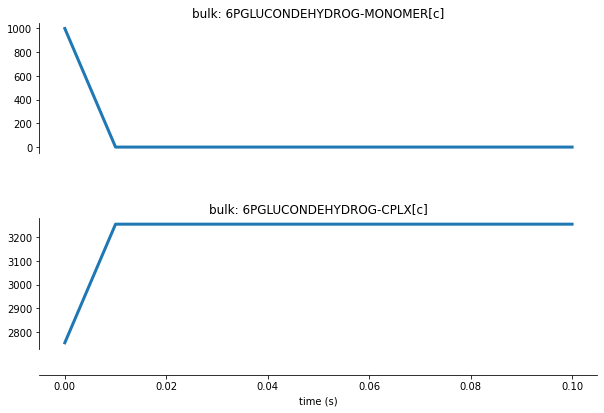
Here we see 6PGLUCONDEHYDROG-MONOMER[c] getting complexed. This a relatively fast process and consumes all the monomers in a single time step.
Transcript Initiation
Transcript Initiation Documentation
[11]:
from ecoli.processes.transcript_initiation import TranscriptInitiation
# load in parameters
ti_params = load_sim_data.get_transcript_initiation_config()
# initialize process and topology
transcript_initiation = TranscriptInitiation(ti_params)
[12]:
# plot topology
ti_topology_plot_settings = {
'node_labels': {
'ecoli-transcript-initiation': 'ecoli\ntranscript\ninitiation',
'full_chromosomes': 'full\nchromosomes',
'listeners\nrna_synth_prob': 'listeners\nrna_synth_\nprob',
'listeners\nribosome_data': 'listeners\nribosome_\ndata',
},
'show_ports': False,
'node_size': 17000,
'node_distance': 3.3,
'dashed_edges': True,
'font_size': 18,
'graph_format': 'hierarchy',
'coordinates': {'ecoli-transcript-initiation': (4, 2)}
}
ti_topology_fig = plot_topology(transcript_initiation, ti_topology_plot_settings)
[13]:
# display ports schema
ti_ports = transcript_initiation.ports_schema()
ti_printout = make_port_printout(ti_ports)
print(ti_printout)
environment:
media_id:
_default:
_updater: set
molecules:
APORNAP-CPLX[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
GUANOSINE-5DP-3DP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
full_chromosomes:
*:
unique_index:
_default: 0
promoters:
*:
TU_index:
_default: 0
coordinates:
_default: 0
domain_index:
_default: 0
bound_TF:
_default: 0
RNAs:
*:
unique_index:
_default: 0
_updater: set
TU_index:
_default: 0
_updater: set
transcript_length:
_default: 0
_updater: set
_emit: True
is_mRNA:
_default: 0
_updater: set
is_full_transcript:
_default: 0
_updater: set
... skipping 2 schema entries ...
active_RNAPs:
*:
unique_index:
_default: 0
_updater: set
domain_index:
_default: 0
_updater: set
coordinates:
_default: 0
_updater: set
_emit: True
direction:
_default: 0
_updater: set
listeners:
mass:
cell_mass:
_default: 0.0
dry_mass:
_default: 0.0
rna_synth_prob:
rna_synth_prob:
_default: 0.0
_updater: set
_emit: True
ribosome_data:
rrn16S_produced:
_default: 0
_updater: set
_emit: True
rrn23S_produced:
_default: 0
_updater: set
_emit: True
rrn5S_produced:
_default: 0
_updater: set
_emit: True
rrn16S_init_prob:
_default: 0
_updater: set
_emit: True
rrn23S_init_prob:
_default: 0
_updater: set
_emit: True
... skipping 2 schema entries ...
rnap_data:
didInitialize:
_default: 0
_updater: set
_emit: True
rnaInitEvent:
_default: 0
_updater: set
_emit: True
[14]:
# run simulation and retrieve final data
ti_settings = {
'total_time': 10,
'initial_state': initial_state,
'topology': transcript_initiation.topology}
ti_data = simulate_process(transcript_initiation, ti_settings)
Simulation ID: a81ba680-2025-11ec-8f9d-8c85908ac627
Created: 09/27/2021 at 23:31:05
Completed in 1.46 seconds
For Transcript Initiation, we can see from the cell above that each active RNA polymerase molecule is represented by an ID number. Let’s analyze how one of these active RNA polymerase molecules functions within this process:
[15]:
# plot output
RNAP_ID = list(ti_data['unique']['active_RNAP'].keys())[0]
ti_fig = plot_variables(
ti_data,
variables=[
('unique', 'active_RNAP', RNAP_ID, 'coordinates'),
('bulk', 'APORNAP-CPLX[c]')
],
column_width=10, row_height=3, row_padding=0.5)
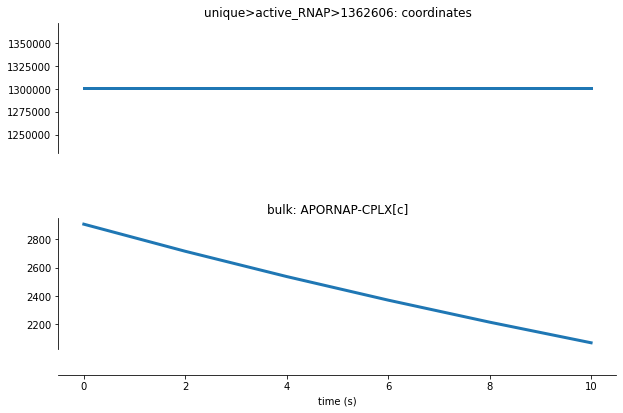
Here we can see that the coordinates for one RNA polymerase molecule is initialized at time=0 and remains the same throughout the simulation as elongation is not a function of this process. Additionally, we can see that the RNA polymerase molecules (given by APORNAP-CPLX[c]) are getting deleted from the bulk molecules count as they bind to the DNA sequence and are initialized for transcription.
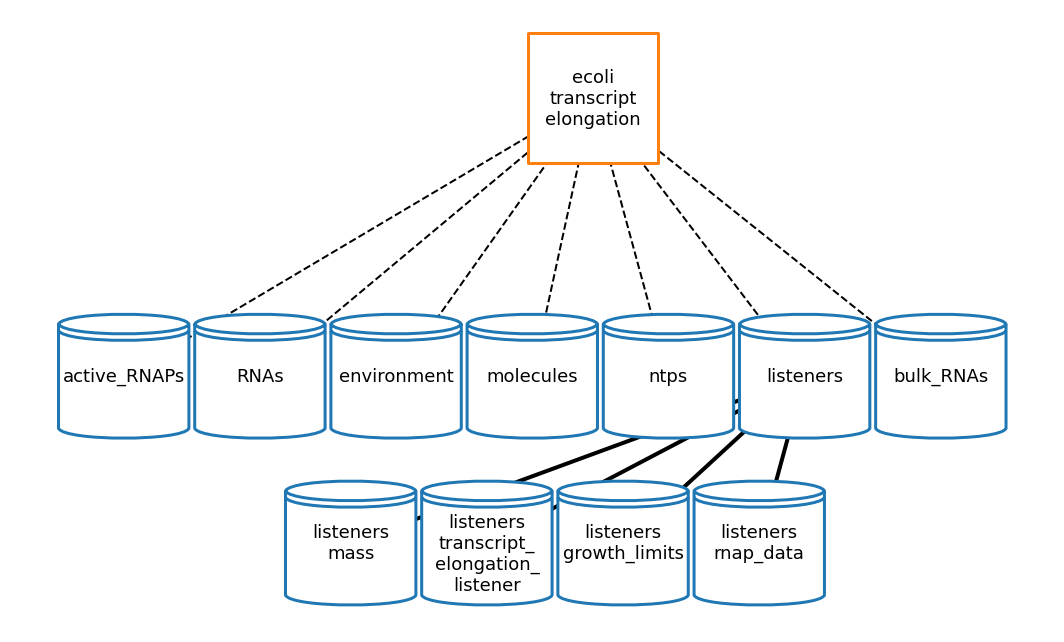
Transcript Elongation
Transcript Elongation Documentation
[16]:
from ecoli.processes.transcript_elongation import TranscriptElongation
# load in parameters
te_params = load_sim_data.get_transcript_elongation_config()
# initialize process and topology
transcript_elongation = TranscriptElongation(te_params)
[17]:
# plot topology
te_topology_plot_settings = {
'node_labels': {
'ecoli-transcript-elongation': 'ecoli\ntranscript\nelongation',
'listeners\ntranscript_elongation_listener': '\nlisteners\ntranscript_\nelongation_\nlistener'
},
'show_ports': False,
'node_size': 17000,
'node_distance': 3.3,
'dashed_edges': True,
'font_size': 18,
'graph_format': 'hierarchy',
'coordinates': {'ecoli-transcript-elongation': (4, 2)}
}
te_topology_fig = plot_topology(transcript_elongation, te_topology_plot_settings)
[18]:
# display ports schema
te_ports = transcript_elongation.ports_schema()
te_printout = make_port_printout(te_ports)
print(te_printout)
environment:
media_id:
_default:
RNAs:
_divider:
<function divide_RNAs_by_domain at 0x12dcc89d0>
topology:
('..', 'active_RNAP')
*:
unique_index:
_default: 0
_updater: set
TU_index:
_default: 0
_updater: set
transcript_length:
_default: 0
_updater: set
_emit: True
is_mRNA:
_default: False
_updater: set
is_full_transcript:
_default: False
_updater: set
... skipping 3 schema entries ...
active_RNAPs:
_divider:
<function divide_active_RNAPs_by_domain at 0x12dcc88b0>
*:
unique_index:
_default: 0
_updater: set
domain_index:
_default: 0
_updater: set
coordinates:
_default: 0
_updater: set
_emit: True
direction:
_default: 0
_updater: set
bulk_RNAs:
EG10001_RNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
EG10002_RNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
EG10003_RNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
EG10004_RNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
EG10006_RNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
... skipping 4682 schema entries ...
ntps:
ATP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
CTP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
GTP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
UTP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
molecules:
PPI[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
APORNAP-CPLX[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
listeners:
mass:
cell_mass:
_default: 0.0
transcript_elongation_listener:
countNTPsUsed:
_default: 0
_updater: set
_emit: True
countRnaSynthesized:
_default: [0. 0. 0. ... 0. 0. 0.]
_updater: set
_emit: True
attenuation_probability:
_updater: set
_emit: True
counts_attenuated:
_updater: set
_emit: True
growth_limits:
ntpUsed:
_default: [0. 0. 0. 0.]
_updater: set
_emit: True
rnap_data:
actualElongations:
_default: 0
_updater: set
_emit: True
didTerminate:
_default: 0
_updater: set
_emit: True
terminationLoss:
_default: 0
_updater: set
_emit: True
didStall:
_updater: set
_emit: True
[19]:
# run simulation and retrieve final data
te_settings = {
'total_time': 10,
'initial_state': initial_state,
'topology': transcript_elongation.topology}
te_data = simulate_process(transcript_elongation, te_settings)
Simulation ID: aa11a2fa-2025-11ec-8f9d-8c85908ac627
Created: 09/27/2021 at 23:31:09
Completed in 2.21 seconds
For Transcript Elongation, we can see from the cell above that each active RNA Polymerase molecule is represented by an ID number. Let’s analyze how a few of these active RNA Polymerase molecules function within this process:
[20]:
# plot output
te_fig = plot_variables(
te_data,
variables=[
('unique', 'active_RNAP', '1266660', 'coordinates'),
('unique', 'active_RNAP', '1293463', 'coordinates'),
('unique', 'active_RNAP', '1293466', 'coordinates')
],
column_width=10, row_height=3, row_padding=0.5)
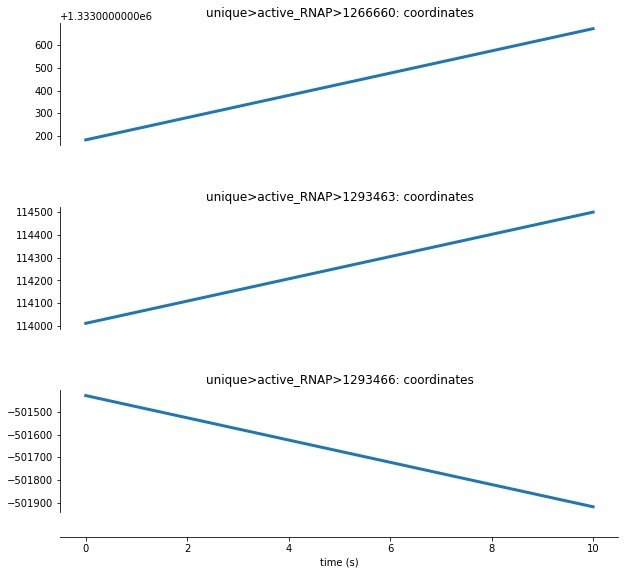
Here we can see that the coordinates for these RNA Polymerase molecules are initialized at time=0 but change throughout the simulation. Some polymerase coordinates incease, indicating elongation in one direction, and others decrease, indicating elongation in the opposite direction along the DNA sequence.
Funadamentally, these changes represent the process of polymerization: as the RNA polymerase molecules travel across the DNA, RNA molecucles are assembled.
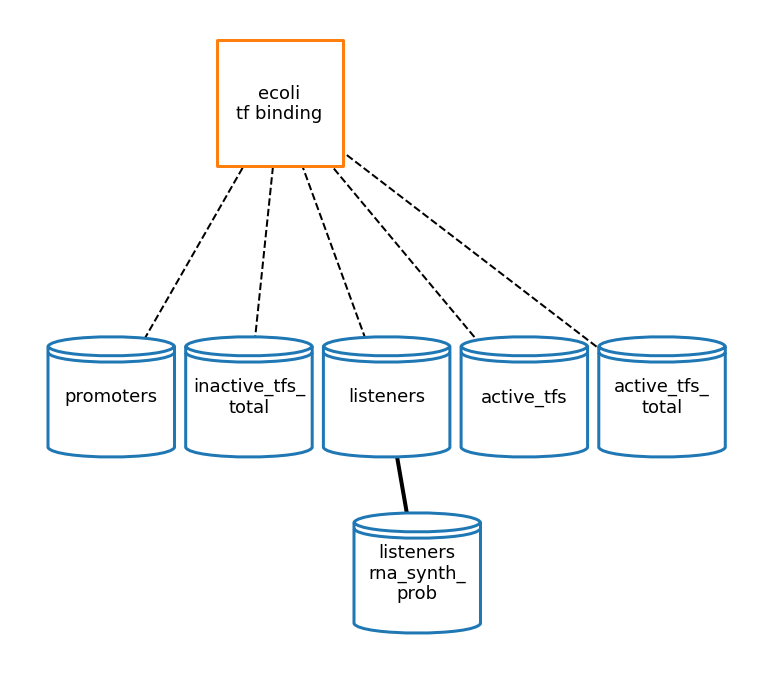
Transcription Factor Binding
[21]:
from ecoli.processes.tf_binding import TfBinding
# load in parameters
tfb_params = load_sim_data.get_tf_config()
# initialize process and topology
tf_binding = TfBinding(tfb_params)
[22]:
# plot topology
tfb_topology_plot_settings = {
'node_labels': {
'ecoli-tf-binding': 'ecoli\ntf binding',
'listeners\nrna_synth_prob': 'listeners\nrna_synth_\nprob',
'active_tfs_total': 'active_tfs_\ntotal',
'inactive_tfs_total': 'inactive_tfs_\ntotal'
},
'show_ports': False,
'node_size': 16000,
'node_distance': 3.5,
'dashed_edges': True,
'font_size': 18,
'graph_format': 'hierarchy',
'coordinates': {'ecoli-tf-binding': (2, 2)}
}
tfb_topology_fig = plot_topology(tf_binding, tfb_topology_plot_settings)
[23]:
# display ports schema
tfb_ports = tf_binding.ports_schema()
tfb_printout = make_port_printout(tfb_ports)
print(tfb_printout)
promoters:
*:
TU_index:
_default: 0
_updater: set
_emit: True
bound_TF:
_default: 0
_updater: set
_emit: True
submass:
_default: [0. 0. 0. 0. 0. 0. 0. 0. 0.]
_emit: True
active_tfs:
CPLX-125[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
CPLX-172[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
CPLX0-226[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
CPLX0-228[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
CPLX0-7669[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
... skipping 19 schema entries ...
active_tfs_total:
CPLX-125[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
CPLX-172[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
CPLX0-226[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
CPLX0-228[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
CPLX0-7669[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
... skipping 19 schema entries ...
inactive_tfs_total:
PC00007[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
PC00003[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
PC00004[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
PC00005[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
CPLX0-7670[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
... skipping 12 schema entries ...
listeners:
rna_synth_prob:
pPromoterBound:
_default: 0
_updater: set
_emit: True
nPromoterBound:
_default: 0
_updater: set
_emit: True
nActualBound:
_default: 0
_updater: set
_emit: True
n_available_promoters:
_default: 0
_updater: set
_emit: True
n_bound_TF_per_TU:
_default: 0
_updater: set
_emit: True
... skipping 1 schema entries ...
[24]:
# run simulation and retrieve final data
tfb_settings = {
'total_time': 10,
'initial_state': initial_state,
'topology': tf_binding.topology}
tfb_data = simulate_process(tf_binding, tfb_settings)
Simulation ID: acb463e4-2025-11ec-8f9d-8c85908ac627
Created: 09/27/2021 at 23:31:12
Completed in 2.07 seconds
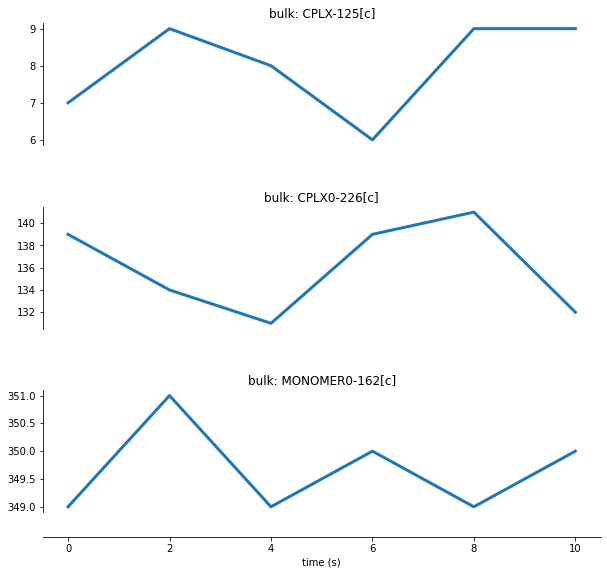
[25]:
# plot output
tfb_fig_active = plot_variables(
tfb_data,
variables=[
('bulk', 'CPLX-125[c]'),
('bulk', 'CPLX0-226[c]'),
('bulk', 'MONOMER0-162[c]')
],
column_width=10, row_height=3, row_padding=0.5)
Chromosome Replication
Chromosome Replication Documentation
[26]:
from ecoli.processes.chromosome_replication import ChromosomeReplication
# load in parameters
cr_params = load_sim_data.get_chromosome_replication_config()
# initialize process and topology
chromosome_replication = ChromosomeReplication(cr_params)
[27]:
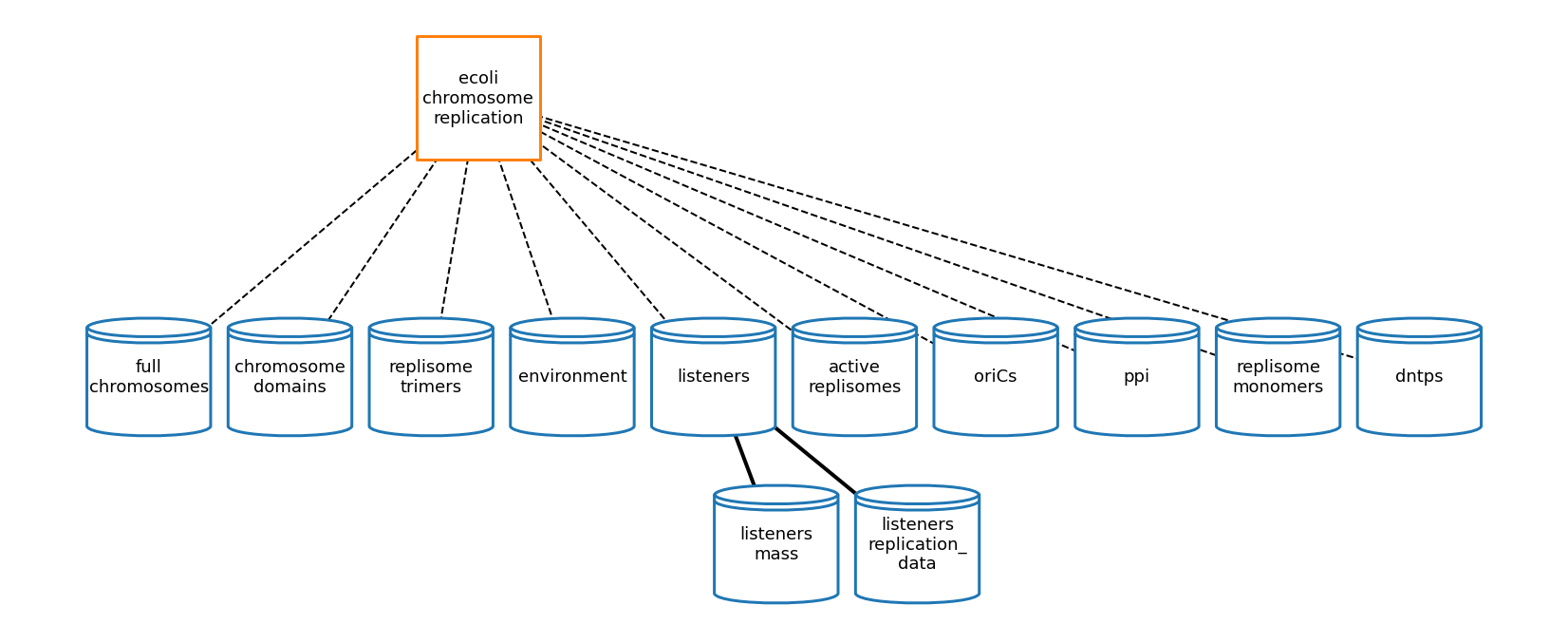
# plot topology
cr_topology_plot_settings = {
'node_labels': {
'ecoli-chromosome-replication': 'ecoli\nchromosome\nreplication',
'replisome_trimers': 'replisome\ntrimers',
'replisome_monomers': 'replisome\nmonomers',
'active_replisomes': 'active\nreplisomes',
'full_chromosomes': 'full\nchromosomes',
'chromosome_domains': 'chromosome\ndomains',
'listeners\nreplication_data': 'listeners\nreplication_\ndata'
},
'show_ports': False,
'node_size': 17000,
'node_distance': 3.5,
'dashed_edges': True,
'font_size': 18,
'graph_format': 'hierarchy',
'coordinates': {'ecoli-chromosome-replication': (3, 2)}
}
cr_topology_fig = plot_topology(chromosome_replication, cr_topology_plot_settings)
[28]:
# display ports schema
cr_ports = chromosome_replication.ports_schema()
cr_printout = make_port_printout(cr_ports)
print(cr_printout)
replisome_trimers:
CPLX0-2361[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
CPLX0-3761[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
replisome_monomers:
CPLX0-3621[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
EG10239-MONOMER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
EG11500-MONOMER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
EG11412-MONOMER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
dntps:
DATP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
DCTP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
DGTP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
TTP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
ppi:
PPI[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
listeners:
mass:
cell_mass:
_default: 0.0
_emit: True
replication_data:
criticalInitiationMass:
_default: 0.0
criticalMassPerOriC:
_default: 0.0
environment:
media_id:
_default:
_updater: set
active_replisomes:
*:
domain_index:
_default: 0
_updater: set
_emit: True
right_replichore:
_default: 0
_updater: set
_emit: True
coordinates:
_default: 0
_updater: set
_emit: True
submass:
_default: [0. 0. 0. 0. 0. 0. 0. 0. 0.]
_emit: True
oriCs:
*:
domain_index:
_default: 0
_updater: set
_emit: True
chromosome_domains:
*:
domain_index:
_default: 0
_updater: set
_emit: True
child_domains:
_default: 0
_updater: set
_emit: True
full_chromosomes:
*:
domain_index:
_default: 0
_updater: set
_emit: True
division_time:
_default: 0
_updater: set
_emit: True
has_triggered_division:
_default: False
_updater: set
[29]:
# tweak initial state to trigger replication
cr_initial_state = copy.deepcopy(initial_state)
cr_initial_state['listeners']['mass']['cell_mass'] = 2000.0
# run simulation and retrieve final data
cr_settings = {
'total_time': 100,
'initial_state': cr_initial_state,
'topology': chromosome_replication.topology,
'emit_step': 10,
'return_raw_data': True}
cr_data = simulate_process(chromosome_replication, cr_settings)
Simulation ID: afd58b16-2025-11ec-8f9d-8c85908ac627
Created: 09/27/2021 at 23:31:17
Completed in 0.349993 seconds
[30]:
cr_data[0.0]['unique']['oriC']
[30]:
{'1': {'domain_index': 1}, '2': {'domain_index': 2}}
[31]:
cr_data[10.0]['unique']['oriC']
[31]:
{'afed02fa-2025-11ec-8f9d-8c85908ac627': {'domain_index': 19},
'afed0390-2025-11ec-8f9d-8c85908ac627': {'domain_index': 20}}
Here a new origin of replication (oriC) has formed between time 0 and 10. This indicates the beginning of the chromosome replication process.
Chromosome Structure
Chromosome Structure Documentation
[32]:
from ecoli.processes.chromosome_structure import ChromosomeStructure
# load in parameters
cs_params = load_sim_data.get_chromosome_structure_config()
# initialize process and topology
chromosome_structure = ChromosomeStructure(cs_params)
[33]:
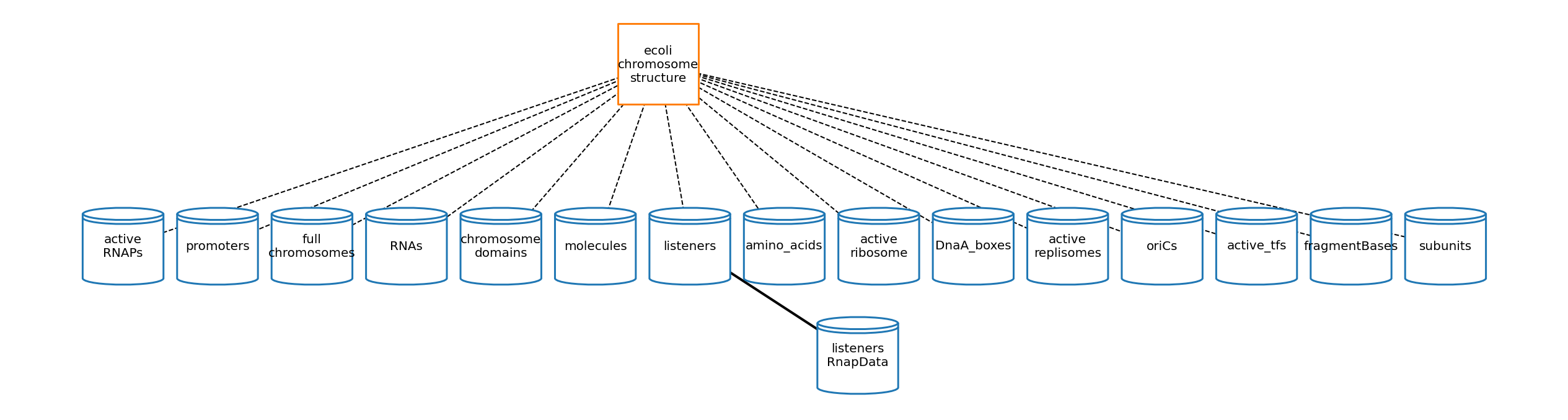
# plot topology
cs_topology_plot_settings = {
'node_labels': {
'ecoli-chromosome-structure': 'ecoli\nchromosome\nstructure',
'full_chromosomes': 'full\nchromosomes',
'active_ribosome': 'active\nribosome',
'chromosome_domains': 'chromosome\ndomains',
'active_replisomes': 'active\nreplisomes',
'active_RNAPs': 'active\nRNAPs',
},
'show_ports': False,
'node_size': 17000,
'node_distance': 3.5,
'dashed_edges': True,
'font_size': 20,
'graph_format': 'hierarchy',
'coordinates': {'ecoli-chromosome-structure': (6, 2)}
}
cs_topology_fig = plot_topology(chromosome_structure, cs_topology_plot_settings)
[34]:
# display ports schema
cs_ports = chromosome_structure.ports_schema()
cs_printout = make_port_printout(cs_ports)
print(cs_printout)
listeners:
RnapData:
n_total_collisions:
_default: 0
_updater: set
_emit: True
n_headon_collisions:
_default: 0
_updater: set
_emit: True
n_codirectional_collisions:
_default: 0
_updater: set
_emit: True
headon_collision_coordinates:
_default: 0
_updater: set
_emit: True
codirectional_collision_coordinates:
_default: 0
_updater: set
_emit: True
... skipping 1 schema entries ...
fragmentBases:
polymerized_ATP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
polymerized_CTP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
polymerized_GTP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
polymerized_UTP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
molecules:
PPI[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
WATER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
APORNAP-CPLX[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
active_tfs:
CPLX-125[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
CPLX-172[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
CPLX0-226[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
CPLX0-228[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
CPLX0-7669[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
... skipping 19 schema entries ...
subunits:
CPLX0-3953[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
CPLX0-3962[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
amino_acids:
L-ALPHA-ALANINE[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
ARG[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
ASN[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
L-ASPARTATE[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
CYS[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
... skipping 16 schema entries ...
active_replisomes:
*:
domain_index:
_default: 0
_updater: set
_emit: True
coordinates:
_default: 0
_updater: set
_emit: True
unique_index:
_default: 0
_updater: set
_emit: True
oriCs:
*:
domain_index:
_default: 0
_updater: set
_emit: True
chromosome_domains:
*:
domain_index:
_default: 0
_updater: set
_emit: True
child_domains:
_default: 0
_updater: set
_emit: True
active_RNAPs:
_divider:
<function divide_active_RNAPs_by_domain at 0x12dcc88b0>
*:
unique_index:
_default: 0
_updater: set
domain_index:
_default: 0
_updater: set
coordinates:
_default: 0
_updater: set
direction:
_default: 0
_updater: set
RNAs:
*:
unique_index:
_default: 0
_updater: set
TU_index:
_default: 0
_updater: set
transcript_length:
_default: 0
_updater: set
RNAP_index:
_default: 0
_updater: set
active_ribosome:
*:
protein_index:
_default: 0
peptide_length:
_default: 0
mRNA_index:
_default: 0
full_chromosomes:
*:
domain_index:
_default: 0
_updater: set
_emit: True
promoters:
*:
TU_index:
_default: 0
coordinates:
_default: 0
domain_index:
_default: 0
bound_TF:
_default: 0
DnaA_boxes:
*:
domain_index:
_default: 0
coordinates:
_default: 0
DnaA_bound:
_default: 0
[35]:
cs_initial_state = copy.deepcopy(initial_state)
# run simulation and retrieve final data
cs_settings = {
'total_time': 10,
'initial_state': cs_initial_state,
'topology': chromosome_structure.topology,
'emit_step': 10,
'return_raw_data': True}
cs_data = simulate_process(chromosome_structure, cs_settings)
Simulation ID: b0a8560e-2025-11ec-8f9d-8c85908ac627
Created: 09/27/2021 at 23:31:20
Completed in 2.53 seconds
Polypeptide Initiation
Polypeptide Initiation Documentation
[36]:
from ecoli.processes.polypeptide_initiation import PolypeptideInitiation
# load in parameters
pi_params = load_sim_data.get_polypeptide_initiation_config()
# initialize process and topology
polypeptide_initiation = PolypeptideInitiation(pi_params)
[37]:
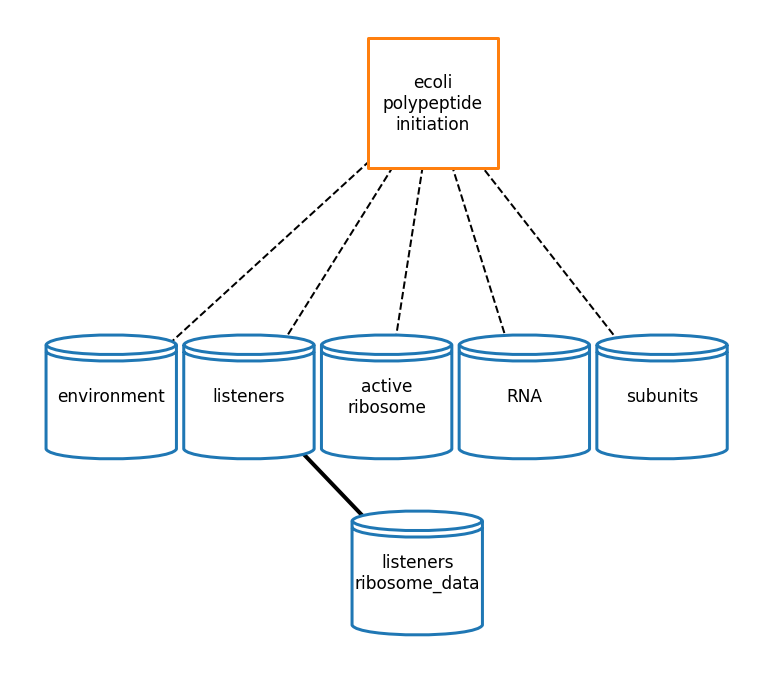
# plot topology
pi_topology_plot_settings = {
'node_labels': {
'ecoli-polypeptide-initiation': 'ecoli\npolypeptide\ninitiation',
'active_ribosome': 'active\nribosome'
},
'show_ports': False,
'node_size': 17000,
'node_distance': 3.5,
'dashed_edges': True,
'font_size': 17,
'graph_format': 'hierarchy',
'coordinates': {'ecoli-polypeptide-initiation': (3, 2)}
}
pi_topology_fig = plot_topology(polypeptide_initiation, pi_topology_plot_settings)
[38]:
# display ports schema
pi_ports = polypeptide_initiation.ports_schema()
pi_printout = make_port_printout(pi_ports)
print(pi_printout)
environment:
media_id:
_default:
_updater: set
listeners:
ribosome_data:
ribosomes_initialized:
_default: 0
_updater: set
_emit: True
prob_translation_per_transcript:
_default: []
_updater: set
_emit: True
effective_elongation_rate:
_default: 0.0
_updater: set
_emit: True
active_ribosome:
*:
unique_index:
_default: 0
protein_index:
_default: 0
peptide_length:
_default: 0
_emit: True
mRNA_index:
_default: 0
pos_on_mRNA:
_default: 0
_emit: True
RNA:
*:
TU_index:
_default: 0
can_translate:
_default: False
unique_index:
_default: 0
subunits:
CPLX0-3953[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
CPLX0-3962[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
[39]:
# run simulation and retrieve final data
pi_settings = {
'total_time': 10,
'initial_state': initial_state,
'topology': polypeptide_initiation.topology}
pi_data = simulate_process(polypeptide_initiation, pi_settings)
Simulation ID: b33343ac-2025-11ec-8f9d-8c85908ac627
Created: 09/27/2021 at 23:31:24
Completed in 2.11 seconds
We can observe the 30S ribosomal subunit (‘CPLX0-3953[c]’) and 50S ribosomal subunit (CPLX0-3962[c]) molecule counts:
[40]:
# plot output
pi_fig = plot_variables(
pi_data,
variables=[
('bulk', 'CPLX0-3953[c]'),
('bulk', 'CPLX0-3962[c]')
],
column_width=10, row_height=3, row_padding=0.5)
pi_fig.savefig('notebooks/pi_fig.png')
pi_fig.show()
/var/folders/t4/q28s90qs5r755xvnzjg6vkc40000gn/T/ipykernel_14901/3946241941.py:11: UserWarning: Matplotlib is currently using module://matplotlib_inline.backend_inline, which is a non-GUI backend, so cannot show the figure.
pi_fig.show()
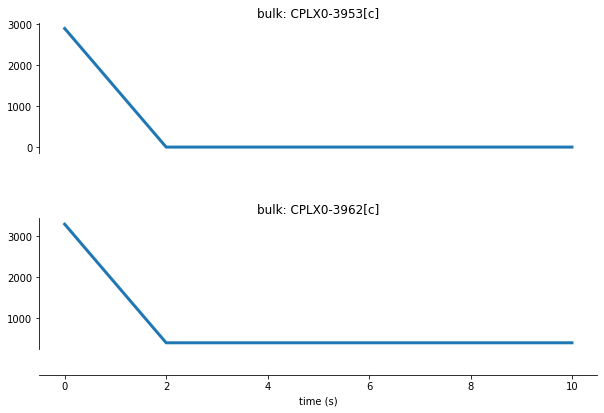
The decrease within the first time step of the simulation demonstrates how active 70S ribosomes are rapidly formed from free 30S and 50S subunits. We can also see how the 30S ribosome subunit (CPLX0-3953[c]) is limiting.
Polypeptide Elongation
Polypeptide Elongation Documentation
[41]:
from ecoli.processes.polypeptide_elongation import PolypeptideElongation
# load in parameters
pe_params = load_sim_data.get_polypeptide_elongation_config()
# initialize process and topology
polypeptide_elongation = PolypeptideElongation(pe_params)
[42]:
# plot topology
pe_topology_plot_settings = {
'node_labels': {
'ecoli-polypeptide-elongation': 'ecoli\npolypeptide\nelongation',
'uncharged_trna': 'uncharged_\ntrna',
'uncharged_trna_total': 'uncharged_\ntrna_total',
'charged_trna_total': 'charged_\ntrna_total',
'charging_molecules': 'charging_\nmolecules',
'active_ribosome': 'active_\nribosome',
'polypeptide_elongation': 'polypeptide\nelongation',
'ppgpp_reaction_metabolites': 'ppgpp\nreaction\nmetabolites',
'chromosome_domains': 'chromosome\ndomains',
'listeners\nreplication_data': 'listeners\nreplication_\ndata',
'amino_acids_total': 'amino_acids_\ntotal'
},
'show_ports': False,
'node_size': 17000,
'node_distance': 3.3,
'dashed_edges': True,
'font_size': 17,
'graph_format': 'hierarchy',
'coordinates': {'ecoli-polypeptide-elongation': (7, 1.75)}
}
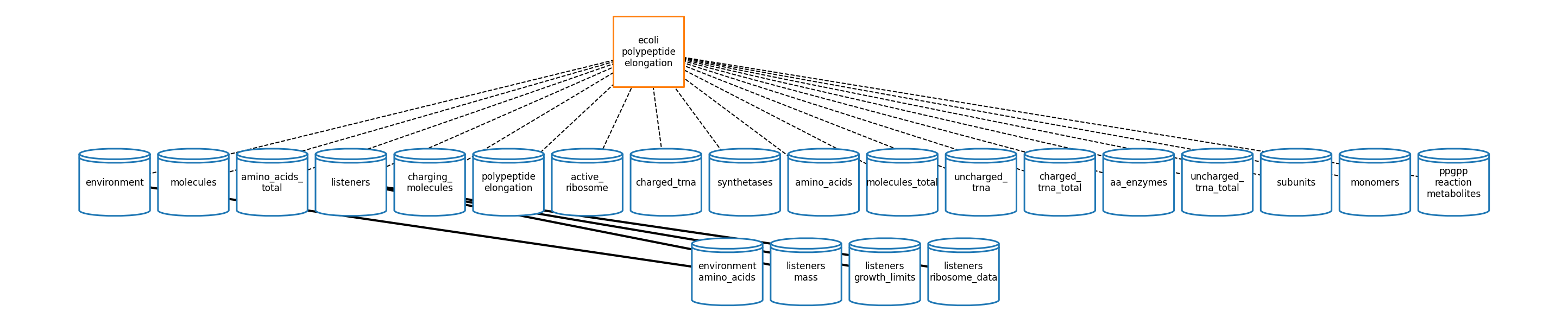
pe_topology_fig = plot_topology(polypeptide_elongation, pe_topology_plot_settings)
[43]:
# display ports schema
pe_ports = polypeptide_elongation.ports_schema()
pe_printout = make_port_printout(pe_ports)
print(pe_printout)
environment:
media_id:
_default:
_updater: set
amino_acids:
L-ALPHA-ALANINE:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
ARG:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
ASN:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
L-ASPARTATE:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
CYS:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
... skipping 16 schema entries ...
listeners:
mass:
cell_mass:
_default: 0.0
dry_mass:
_default: 0.0
growth_limits:
fraction_trna_charged:
_default: 0
_updater: set
_emit: True
aa_pool_size:
_default: 0
_updater: set
_emit: True
aa_request_size:
_default: 0
_updater: set
_emit: True
active_ribosomes_allocated:
_default: 0
_updater: set
_emit: True
net_charged:
_default: []
_updater: set
_emit: True
... skipping 5 schema entries ...
ribosome_data:
translation_supply:
_default: 0
_updater: set
_emit: True
effective_elongation_rate:
_default: 0
_updater: set
_emit: True
aaCountInSequence:
_default: 0
_updater: set
_emit: True
aaCounts:
_default: 0
_updater: set
_emit: True
actualElongations:
_default: 0
_updater: set
_emit: True
... skipping 6 schema entries ...
molecules:
PROTON[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
WATER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
RELA-MONOMER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
SPOT-MONOMER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
GUANOSINE-5DP-3DP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
molecules_total:
PROTON[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
WATER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
RELA-MONOMER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
SPOT-MONOMER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
GUANOSINE-5DP-3DP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
monomers:
1-ACYLGLYCEROL-3-P-ACYLTRANSFER-MONOMER[i]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
1-PFK-MONOMER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
2-DEHYDROPANTOATE-REDUCT-MONOMER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
2-ISOPROPYLMALATESYN-MONOMER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
2-OCTAPRENYL-METHOXY-BENZOQ-METH-MONOMER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
... skipping 4291 schema entries ...
amino_acids:
L-ALPHA-ALANINE[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
ARG[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
ASN[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
L-ASPARTATE[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
CYS[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
... skipping 16 schema entries ...
aa_enzymes:
2-ISOPROPYLMALATESYN-MONOMER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
ACETOLACTSYNI-CPLX[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
ANTHRANSYN-CPLX[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
ASNSYNA-CPLX[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
ASPAMINOTRANS-DIMER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
... skipping 21 schema entries ...
ppgpp_reaction_metabolites:
ATP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
GDP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
AMP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
GUANOSINE-5DP-3DP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
PROTON[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
... skipping 2 schema entries ...
uncharged_trna:
alaT-tRNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
alaU-tRNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
alaV-tRNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
alaW-tRNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
alaX-tRNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
... skipping 81 schema entries ...
charged_trna:
charged-alaT-tRNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
charged-alaU-tRNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
charged-alaV-tRNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
charged-alaW-tRNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
charged-alaX-tRNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
... skipping 81 schema entries ...
charging_molecules:
L-ALPHA-ALANINE[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
ATP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
alaT-tRNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
PPI[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
AMP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
... skipping 190 schema entries ...
synthetases:
ALAS-CPLX[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
ARGS-MONOMER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
ASNS-CPLX[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
ASPS-CPLX[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
CYSS-MONOMER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
... skipping 16 schema entries ...
amino_acids_total:
L-ALPHA-ALANINE[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
ARG[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
ASN[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
L-ASPARTATE[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
CYS[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
... skipping 16 schema entries ...
uncharged_trna_total:
alaT-tRNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
alaU-tRNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
alaV-tRNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
alaW-tRNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
alaX-tRNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
... skipping 81 schema entries ...
charged_trna_total:
charged-alaT-tRNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
charged-alaU-tRNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
charged-alaV-tRNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
charged-alaW-tRNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
charged-alaX-tRNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
... skipping 81 schema entries ...
active_ribosome:
*:
unique_index:
_default: 0
_updater: set
protein_index:
_default: 0
_updater: set
peptide_length:
_default: 0
_updater: set
_emit: True
pos_on_mRNA:
_default: 0
_updater: set
_emit: True
submass:
_default: [0. 0. 0. 0. 0. 0. 0. 0. 0.]
_emit: True
subunits:
CPLX0-3953[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
CPLX0-3962[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
polypeptide_elongation:
aa_count_diff:
_default: {}
_updater: set
_emit: True
gtp_to_hydrolyze:
_default: 0
_updater: set
_emit: True
[44]:
# run simulation and retrieve final data
pe_settings = {
'total_time': 10,
'initial_state': initial_state,
'topology': polypeptide_elongation.topology}
pe_data = simulate_process(polypeptide_elongation, pe_settings)
Simulation ID: b61fa8a8-2025-11ec-8f9d-8c85908ac627
Created: 09/27/2021 at 23:31:29
Completed in 3.24 seconds
We can see from the cell above that each active ribosome molecule is represented by an ID number. Let’s analyze the polypeptide length and the ribosome’s position on mRNA of one active ribosome within this process:
[45]:
# plot output
RIBOSOME_ID = '1246839'
pe_fig = plot_variables(
pe_data,
variables=[
('unique', 'active_ribosome', RIBOSOME_ID, 'pos_on_mRNA'),
('unique', 'active_ribosome', RIBOSOME_ID, 'peptide_length')
],
column_width=10, row_height=3, row_padding=0.5)
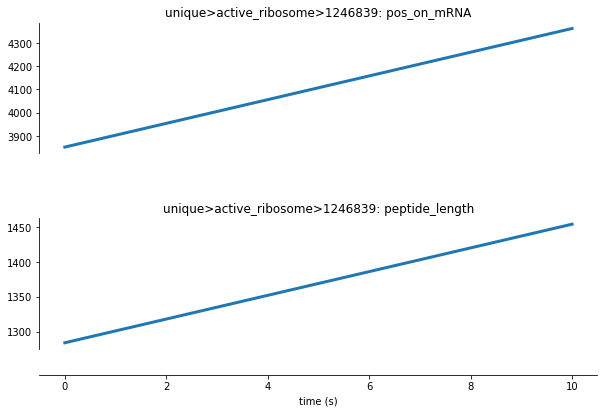
Here we can see that as the simulation progresses, the ribosome travels along the mRNA strand (as shown by the increasing pos_on_mRNA variable) and polymerization of amino acids into a polypeptide occurs (as shown by the increasing peptide_length variable).
After elongation terminates, we can see an increase in protein counts:
[46]:
pe_fig1 = plot_variables(
pe_data,
variables=[
('bulk', '6PGLUCONDEHYDROG-MONOMER[c]')
],
column_width=10, row_height=3, row_padding=0.5)
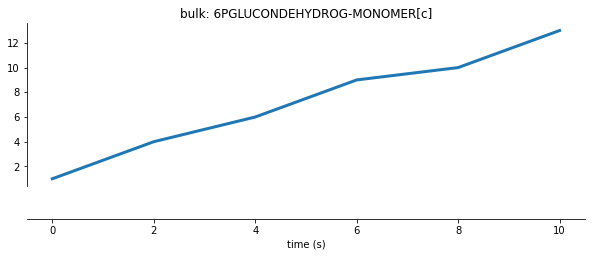
Protein Degradation
Polypeptide Degradation Documentation
[47]:
from ecoli.processes.protein_degradation import ProteinDegradation
# load in parameters
pd_params = load_sim_data.get_protein_degradation_config()
# initialize process and topology
protein_degradation = ProteinDegradation(pd_params)
[48]:
# plot topology
pd_topology_plot_settings = {
'buffer': 1,
'node_labels': {
'ecoli-protein-degradation': 'ecoli\nprotein\ndegradation'
},
'node_distance': 5,
'show_ports': False,
'node_size': 10000,
'dashed_edges': True,
'coordinates': {'ecoli-protein-degradation': (1.5, 0.5)}
}
pd_topology_fig = plot_topology(protein_degradation, pd_topology_plot_settings)
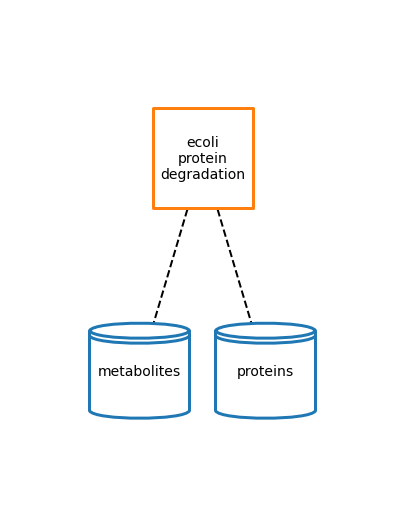
[49]:
# display ports schema
pd_ports = protein_degradation.ports_schema()
pd_printout = make_port_printout(pd_ports)
print(pd_printout)
metabolites:
L-ALPHA-ALANINE[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
ARG[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
ASN[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
L-ASPARTATE[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
CYS[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
... skipping 17 schema entries ...
proteins:
1-ACYLGLYCEROL-3-P-ACYLTRANSFER-MONOMER[i]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
1-PFK-MONOMER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
2-DEHYDROPANTOATE-REDUCT-MONOMER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
2-ISOPROPYLMALATESYN-MONOMER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
2-OCTAPRENYL-METHOXY-BENZOQ-METH-MONOMER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
... skipping 4291 schema entries ...
[50]:
# run simulation and retrieve final data
pd_settings = {
'total_time': 600,
'initial_state': initial_state,
'topology': protein_degradation.topology,
'emit_step': 10}
pd_data = simulate_process(protein_degradation, pd_settings)
Simulation ID: ba09488e-2025-11ec-8f9d-8c85908ac627
Created: 09/27/2021 at 23:31:34
Completed in 5.69 seconds
For protein degradation, let’s look at the ARTJ-MONOMER[p] monomer, a protein selected for degradation at this time point, as it degrades into its subcomponents:
[51]:
# plot output
pd_fig = plot_variables(
pd_data,
variables=[
('bulk', 'ARTJ-MONOMER[p]'),
('bulk', 'ASN[c]'),
('bulk', 'WATER[c]')
],
column_width=10, row_height=3, row_padding=0.5)
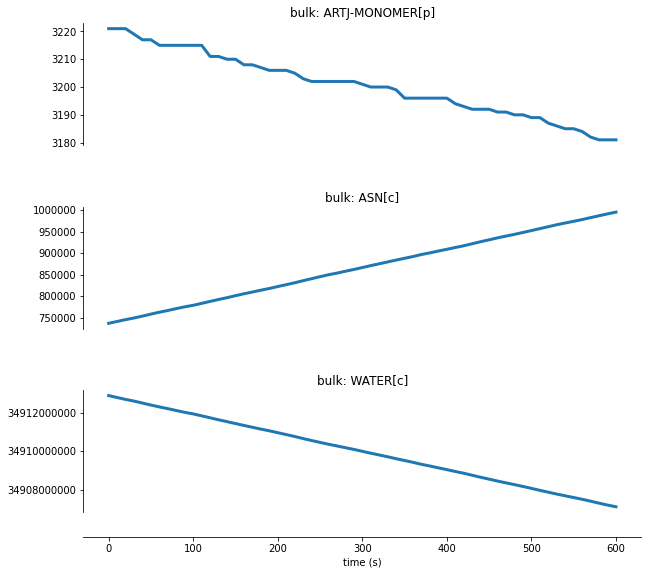
We can see here that as the ARTJ-MONOMER[p] protein degrades, the count for the amino acid asparagine increases and water is consumed.
RNA Degradation
[52]:
from ecoli.processes.rna_degradation import RnaDegradation
# load in parameters
rd_params = load_sim_data.get_rna_degradation_config()
# initialize process and topology
rna_degradation = RnaDegradation(rd_params)
[53]:
# plot topology
rd_topology_plot_settings = {
'node_labels': {
'ecoli-rna-degradation': 'ecoli\nrna\ndegradation',
'fragmentMetabolites': 'fragment\nMetabolites',
'listeners\nrna_degradation_listener': '\nlisteners\nrna_\ndegradation_\nlistener',
'active_ribosome': 'active_\nribosome'
},
'show_ports': False,
'node_size': 17000,
'node_distance': 3.3,
'dashed_edges': True,
'font_size': 17,
'graph_format': 'hierarchy',
'coordinates': {'ecoli-rna-degradation': (7, 1.75)}
}
rd_topology_fig = plot_topology(rna_degradation, rd_topology_plot_settings)
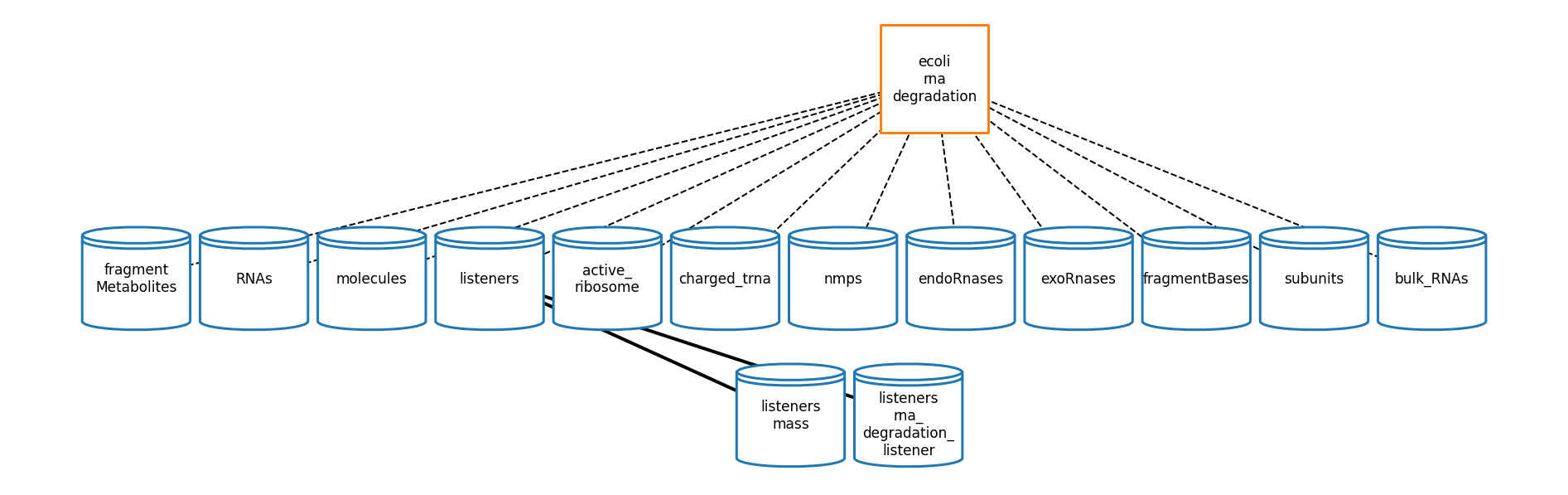
[54]:
# display ports schema
rd_ports = rna_degradation.ports_schema()
rd_printout = make_port_printout(rd_ports)
print(rd_printout)
charged_trna:
charged-alaT-tRNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
charged-alaU-tRNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
charged-alaV-tRNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
charged-alaW-tRNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
charged-alaX-tRNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
... skipping 81 schema entries ...
bulk_RNAs:
EG10001_RNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
_updater: nonnegative_accumulate
EG10002_RNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
_updater: nonnegative_accumulate
EG10003_RNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
_updater: nonnegative_accumulate
EG10004_RNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
_updater: nonnegative_accumulate
EG10006_RNA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
_updater: nonnegative_accumulate
... skipping 4682 schema entries ...
nmps:
AMP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
CMP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
GMP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
UMP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
fragmentMetabolites:
polymerized_ATP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
polymerized_CTP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
polymerized_GTP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
polymerized_UTP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
WATER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
... skipping 2 schema entries ...
fragmentBases:
polymerized_ATP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
polymerized_CTP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
polymerized_GTP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
polymerized_UTP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
endoRnases:
EG10856-MONOMER[p]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
EG10857-MONOMER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
EG10859-MONOMER[i]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
EG10860-MONOMER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
EG10861-MONOMER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
... skipping 4 schema entries ...
exoRnases:
EG11620-MONOMER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
G7175-MONOMER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
EG10858-MONOMER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
EG10863-MONOMER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
EG11259-MONOMER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
... skipping 4 schema entries ...
subunits:
CPLX0-3953[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
CPLX0-3962[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
molecules:
WATER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
PROTON[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
active_ribosome:
*:
unique_index:
_default: 0
RNAs:
*:
unique_index:
_default: 0
_updater: set
TU_index:
_default: 0
_updater: set
is_full_transcript:
_default: 0
_updater: set
can_translate:
_default: 0
_updater: set
listeners:
mass:
cell_mass:
_default: 0.0
dry_mass:
_default: 0.0
rna_degradation_listener:
fraction_active_endo_rnases:
_default: 0
_updater: set
_emit: True
diff_relative_first_order_decay:
_default: 0
_updater: set
_emit: True
fract_endo_rrna_counts:
_default: 0
_updater: set
_emit: True
count_rna_degraded:
_default: 0
_updater: set
_emit: True
nucleotides_from_degradation:
_default: 0
_updater: set
_emit: True
... skipping 1 schema entries ...
[55]:
# run simulation and retrieve final data
rd_settings = {
'total_time': 100,
'initial_state': initial_state,
'topology': rna_degradation.topology}
rd_data = simulate_process(rna_degradation, rd_settings)
Simulation ID: bdf352a0-2025-11ec-8f9d-8c85908ac627
Created: 09/27/2021 at 23:31:42
Completed in 10.27 seconds
For RNA degradation, let’s look at a few RNA molecules as the simulation runs:
[56]:
# plot output
rd_fig = plot_variables(
rd_data,
variables=[
('bulk', 'RNA0-300[c]'),
('bulk', 'RNA0-301[c]'),
('bulk', 'WATER[c]')
],
column_width=10, row_height=3, row_padding=0.5)
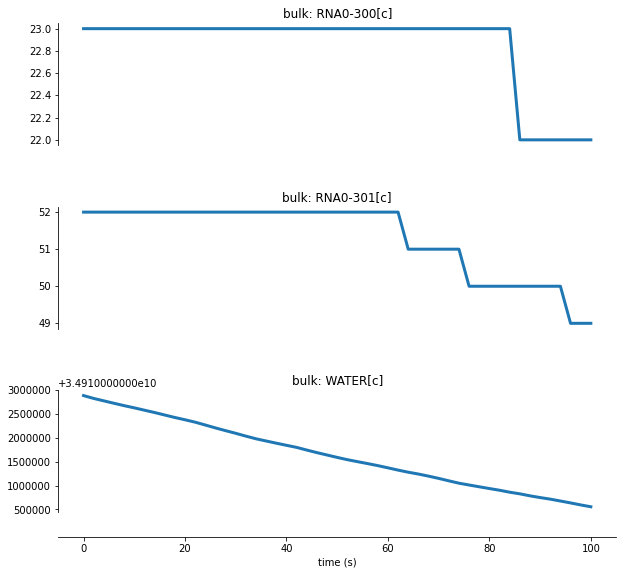
RNAs are selected and degraded by endoRNases, and non-functional RNA fragments are digested through exoRNases. During the process water is consumed, and nucleotides, pyrophosphate and protons are released.
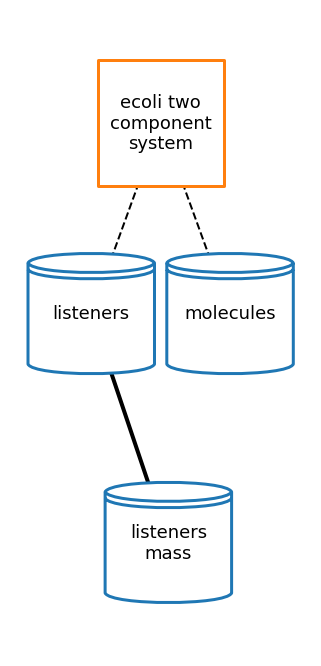
Two Component System
Two Component System Documentation
[57]:
from ecoli.processes.two_component_system import TwoComponentSystem
# load in parameters
tcs_params = load_sim_data.get_two_component_system_config()
# initialize process and topology
two_component_system = TwoComponentSystem(tcs_params)
[58]:
# plot topology
tcs_topology_plot_settings = {
'node_labels': {
'ecoli-two-component-system': 'ecoli two\ncomponent\nsystem'
},
'show_ports': False,
'node_size': 16000,
'node_distance': 5.0,
'dashed_edges': True,
'font_size': 18,
'graph_format': 'hierarchy',
'coordinates': {'ecoli-two-component-system': (1.35, 1)}
}
tcs_topology_fig = plot_topology(two_component_system, tcs_topology_plot_settings)
[59]:
# display ports schema
tcs_ports = two_component_system.ports_schema()
tcs_printout = make_port_printout(tcs_ports)
print(tcs_printout)
molecules:
PHOSPHO-ARCB-CPLX[i]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
ADP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
PROTON[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
ARCB-CPLX[i]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
ATP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
... skipping 36 schema entries ...
listeners:
mass:
cell_mass:
_default: 0
[60]:
# tweak initial state??
tcs_initial_state = copy.deepcopy(initial_state)
# run simulation and retrieve final data
tcs_settings = {
'total_time': 10,
'initial_state': initial_state,
'topology': two_component_system.topology}
tcs_data = simulate_process(two_component_system, tcs_settings)
Simulation ID: c62fb3f0-2025-11ec-8f9d-8c85908ac627
Created: 09/27/2021 at 23:31:55
Completed in 0.038426 seconds
[61]:
tcs_data
[61]:
{'bulk': {'PHOSPHO-ARCB-CPLX[i]': [0, 0, 0, 0, 0, 0],
'ADP[c]': [404873, 404873, 404873, 404873, 404873, 404873],
'PROTON[c]': [52, 52, 52, 52, 52, 52],
'ARCB-CPLX[i]': [0, 0, 0, 0, 0, 0],
'ATP[c]': [7866030, 7866030, 7866030, 7866030, 7866030, 7866030],
'PHOSPHO-ARCA[c]': [0, 0, 0, 0, 0, 0],
'ARCA-MONOMER[c]': [2769, 2769, 2769, 2769, 2769, 2769],
'Pi[c]': [408412, 408412, 408412, 408412, 408412, 408412],
'WATER[c]': [34912886892,
34912886892,
34912886892,
34912886892,
34912886892,
34912886892],
'PHOSPHO-BAES-INDOLE-CPLX[i]': [0, 0, 0, 0, 0, 0],
'BAES-INDOLE-CPLX[i]': [0, 0, 0, 0, 0, 0],
'PHOSPHO-BAER[c]': [0, 0, 0, 0, 0, 0],
'BAER-MONOMER[c]': [200, 200, 200, 200, 200, 200],
'PHOSPHO-BAES[i]': [0, 0, 0, 0, 0, 0],
'BAES-MONOMER[i]': [19, 19, 19, 19, 19, 19],
'PHOSPHO-BASS-FE+3-CPLX[i]': [0, 0, 0, 0, 0, 0],
'BASS-FE+3-CPLX[i]': [0, 0, 0, 0, 0, 0],
'PHOSPHO-BASR[c]': [0, 0, 0, 0, 0, 0],
'BASR-MONOMER[c]': [180, 180, 180, 180, 180, 180],
'PHOSPHO-BASS[i]': [0, 0, 0, 0, 0, 0],
'BASS-MONOMER[i]': [46, 46, 46, 46, 46, 46],
'PHOSPHO-DCUS-SUC-CPLX[i]': [0, 0, 0, 0, 0, 0],
'DCUS-SUC-CPLX[p]': [0, 0, 0, 0, 0, 0],
'PHOSPHO-DCUR[c]': [0, 0, 0, 0, 0, 0],
'DCUR-MONOMER[c]': [80, 80, 80, 80, 80, 80],
'PHOSPHO-DCUS[i]': [0, 0, 0, 0, 0, 0],
'DCUS-MONOMER[p]': [12, 12, 12, 12, 12, 12],
'PHOSPHO-NARX-NITRATE-CPLX[i]': [0, 0, 0, 0, 0, 0],
'NARX-NITRATE-CPLX[i]': [0, 0, 0, 0, 0, 0],
'PHOSPHO-NARL[c]': [0, 0, 0, 0, 0, 0],
'NARL-MONOMER[c]': [679, 679, 679, 679, 679, 679],
'PHOSPHO-NARX[i]': [0, 0, 0, 0, 0, 0],
'NARX-CPLX[i]': [17, 17, 17, 17, 17, 17],
'PHOSPHO-PHOQ[i]': [1, 1, 1, 1, 1, 1],
'CPLX0-8168[i]': [13, 13, 13, 13, 13, 13],
'PHOSPHO-PHOP[c]': [148, 148, 148, 148, 148, 148],
'PHOP-MONOMER[c]': [855, 855, 855, 855, 855, 855],
'PHOSPHO-PHOR[i]': [0, 0, 0, 0, 0, 0],
'PHOR-CPLX[c]': [0, 0, 0, 0, 0, 0],
'PHOSPHO-PHOB[c]': [0, 0, 0, 0, 0, 0],
'PHOB-MONOMER[c]': [88, 88, 88, 88, 88, 88]},
'listeners': {'mass': {}},
'time': [0.0, 2.0, 4.0, 6.0, 8.0, 10.0]}
Phosphate groups are transferred from histidine kinases to response regulators and back in response to counts of ligand stimulants
[62]:
# # plot output
# tcs_fig = plot_variables(
# tcs_data,
# variables=[
# ],
# column_width=10, row_height=3, row_padding=0.5)
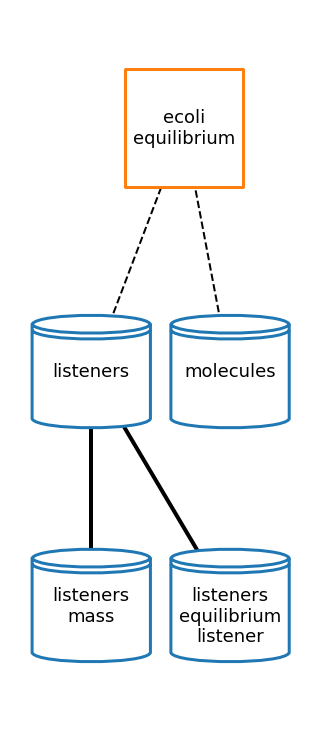
Equilibrium
[63]:
from ecoli.processes.equilibrium import Equilibrium
# load in parameters
eq_params = load_sim_data.get_equilibrium_config()
# initialize process and topology
equilibrium = Equilibrium(eq_params)
[64]:
# plot topology
eq_topology_plot_settings = {
'node_labels': {
'ecoli-equilibrium': 'ecoli\nequilibrium',
'listeners\nequilibrium_listener': '\nlisteners\nequilibrium\nlistener'
},
'show_ports': False,
'node_size': 14000,
'node_distance': 5.0,
'dashed_edges': True,
'font_size': 18,
'graph_format': 'hierarchy',
'coordinates': {'ecoli-equilibrium': (1.5, 1.25)}
}
eq_topology_fig = plot_topology(equilibrium, eq_topology_plot_settings)
[65]:
# display ports schema
eq_ports = equilibrium.ports_schema()
eq_printout = make_port_printout(eq_ports)
print(eq_printout)
molecules:
CPLX-123[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
PC00033[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
HYPOXANTHINE[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
CPLX-125[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
TRP[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
... skipping 93 schema entries ...
listeners:
mass:
cell_mass:
_default: 0
equilibrium_listener:
reaction_rates:
_default: []
_updater: set
_emit: True
[66]:
# run simulation and retrieve final data
eq_settings = {
'total_time': 10,
'initial_state': initial_state,
'topology': equilibrium.topology}
eq_data = simulate_process(equilibrium, eq_settings)
Simulation ID: c64fab9c-2025-11ec-8f9d-8c85908ac627
Created: 09/27/2021 at 23:31:55
Completed in 0.182374 seconds
[67]:
# # plot output
# eq_fig = plot_variables(
# eq_data,
# variables=[
# ],
# column_width=10, row_height=3, row_padding=0.5)
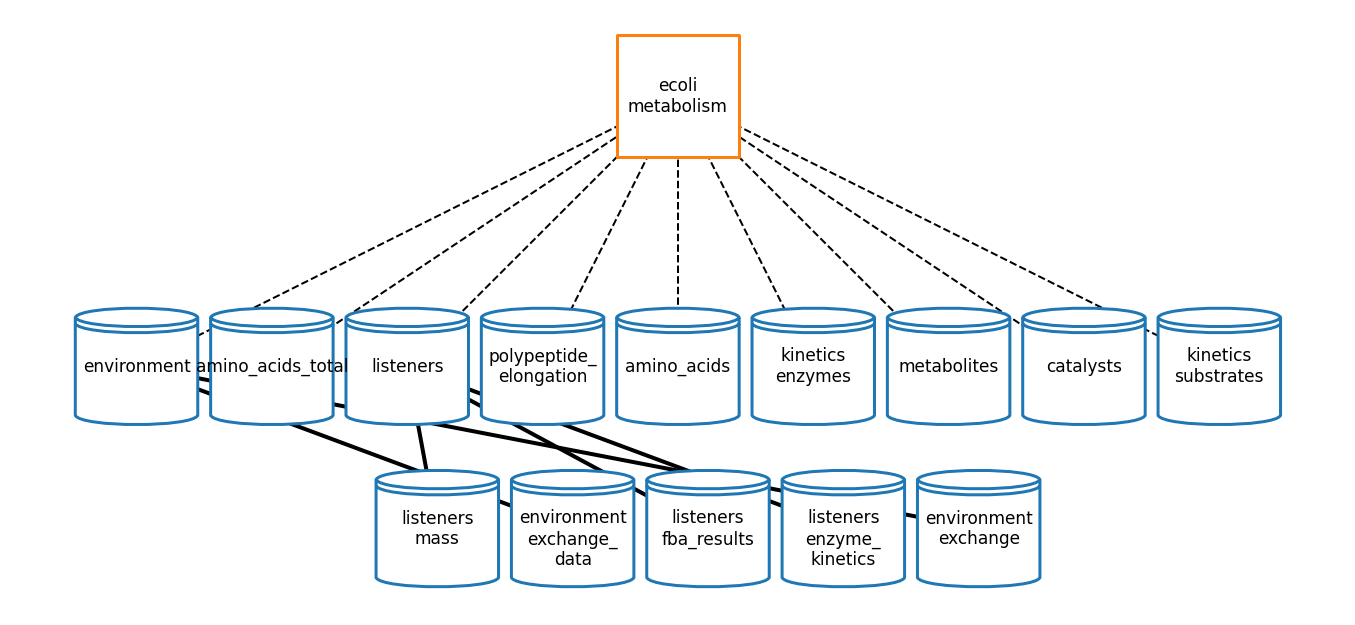
Metabolism
[68]:
from ecoli.processes.metabolism import Metabolism
# load in parameters
meta_params = load_sim_data.get_metabolism_config()
# initialize process and topology
metabolism = Metabolism(meta_params)
[69]:
# plot topology
meta_topology_plot_settings = {
'node_labels': {
'ecoli-metabolism': 'ecoli\nmetabolism',
'kinetics_enzymes': 'kinetics\nenzymes',
'kinetics_substrates': 'kinetics\nsubstrates',
'environment\nexchange_data': '\nenvironment\nexchange_\ndata',
'listeners\nenzyme_kinetics': '\nlisteners\nenzyme_\nkinetics',
'polypeptide_elongation': 'polypeptide_\nelongation'
},
'show_ports': False,
'node_size': 15000,
'node_distance': 3.2,
'dashed_edges': True,
'font_size': 17,
'graph_format': 'hierarchy',
'coordinates': {'ecoli-metabolism': (4.5, 2)}
}
meta_topology_fig = plot_topology(metabolism, meta_topology_plot_settings)
[70]:
# display ports schema
meta_ports = metabolism.ports_schema()
meta_printout = make_port_printout(meta_ports)
print(meta_printout)
metabolites:
2-3-DIHYDROXYBENZOATE[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
2-KETOGLUTARATE[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
2-PG[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
2K-4CH3-PENTANOATE[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
4-AMINO-BUTYRATE[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
... skipping 167 schema entries ...
catalysts:
1-ACYLGLYCEROL-3-P-ACYLTRANSFER-MONOMER[i]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
1-PFK[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
2-DEHYDROPANTOATE-REDUCT-MONOMER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
2-ISOPROPYLMALATESYN-MONOMER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
2-OCTAPRENYL-METHOXY-BENZOQ-METH-MONOMER[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
... skipping 1472 schema entries ...
kinetics_enzymes:
3-ISOPROPYLMALDEHYDROG-CPLX[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
6PFK-1-CPX[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
6PFK-2-CPX[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
7-ALPHA-HYDROXYSTEROID-DEH-CPLX[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
7KAPSYN-CPLX[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
... skipping 307 schema entries ...
kinetics_substrates:
2-3-DIHYDROXYBENZOATE[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
2-KETOGLUTARATE[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
4-AMINO-BUTYRATE[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
4-hydroxybenzoate[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
ACETYL-COA[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
... skipping 86 schema entries ...
amino_acids:
L-ALPHA-ALANINE[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
ARG[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
ASN[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
L-ASPARTATE[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
CYS[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
_properties: {'bulk': True}
... skipping 16 schema entries ...
amino_acids_total:
L-ALPHA-ALANINE[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
ARG[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
ASN[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
L-ASPARTATE[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
CYS[c]:
_default: 0
_divider: binomial_ecoli
_emit: True
... skipping 16 schema entries ...
environment:
media_id:
_default:
_updater: set
exchange:
ACET[p]:
_default: 0
AMMONIUM[c]:
_default: 0
BETAINE[p]:
_default: 0
BUTANAL[c]:
_default: 0
CA+2[p]:
_default: 0
... skipping 46 schema entries ...
exchange_data:
unconstrained:
_default: []
constrained:
_default: []
listeners:
mass:
cell_mass:
_default: 0.0
dry_mass:
_default: 0.0
fba_results:
media_id:
_default:
_updater: set
conc_updates:
_default: []
_updater: set
catalyst_counts:
_default: 0
_updater: set
translation_gtp:
_default: 0
_updater: set
coefficient:
_default: 0.0
_updater: set
... skipping 12 schema entries ...
enzyme_kinetics:
metaboliteCountsInit:
_default: 0
_updater: set
metaboliteCountsFinal:
_default: 0
_updater: set
enzymeCountsInit:
_default: 0
_updater: set
countsToMolar:
_default: 1.0
_updater: set
actualFluxes:
_default: []
_updater: set
... skipping 3 schema entries ...
polypeptide_elongation:
aa_count_diff:
_default: {}
_emit: True
gtp_to_hydrolyze:
_default: 0
_emit: True
[71]:
# run simulation and retrieve final data
meta_settings = {
'total_time': 10,
'initial_state': initial_state,
'topology': metabolism.topology}
meta_data = simulate_process(metabolism, meta_settings)
Simulation ID: c73b86d4-2025-11ec-8f9d-8c85908ac627
Created: 09/27/2021 at 23:31:57
Completed in 1.80 seconds
[72]:
reaction_fluxes = meta_data['listeners']['fba_results']['reactionFluxes']
del reaction_fluxes[0]
[73]:
# generate time series
reaction_fluxes_time_series = {}
for fluxes in reaction_fluxes:
for reaction_id, flux in enumerate(fluxes):
if reaction_id not in reaction_fluxes_time_series:
reaction_fluxes_time_series[reaction_id] = []
reaction_fluxes_time_series[reaction_id].append(flux)
[74]:
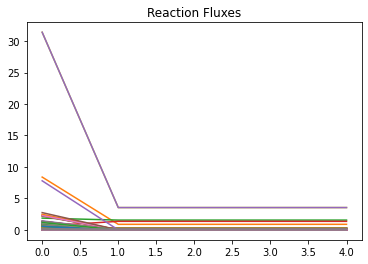
# plot figure
import matplotlib.pyplot as plt
fig = plt.figure()
for reaction_id, time_series in reaction_fluxes_time_series.items():
plt.plot(time_series)
plt.title('Reaction Fluxes')
[74]:
Text(0.5, 1.0, 'Reaction Fluxes')